TENT5A and Nucleic Acid Persistence sheds light on more vaccine slop
Apr 29, 2025

This is the 5th paper where plasmid DNA sequences can be found in patients or mice post vaccination with C19 mRNA vaccines. This particular paper is a gold mine and it does a thorough job of teasing apart unappreciated biology in cells post transfection It also follows up its hypothesis with knock out models and Nanopore sequencing to dissect the problem. There is a staggering amount of work that went into this paper.
There was an active debate and productive discussion with its lead author many years ago when first published its preprint form.
Nepetalactone Newsletter is a reader-supported publication. To receive new posts and support my work, consider becoming a free or paid subscriber.
Why…
There has been a nagging concern over the implications of the poor RIN scores on the modRNA in the vaccines (Blotgate). This is a metric that radically changed when they moved from PCR manufactured vaccines to E.coli manufactured vaccines.

The EMA noted the RIN scores dropped from 75% to 50%. A RIN score is a RNA Integrity Number which is a measure of how degraded the RNA is. Higher scores are more pure full length mRNA lots. Lower scores are fragmented, degraded or off target synthesis.
Are these stalled extension products of the T7 RNA polymerase? Are they fragmented via metal contamination? Are they template switched T7 polymerase products? What are their clinical implications with such an impure prodrug? These are prodrugs as the mRNA is translated into a theoretical spike protein and that translation process likely varies in every cell and person injected. This biodistribution is known to be more expansive than the virus.
The paper title changed from PrePrint to Nature Publication

You’ll note it changed from ‘enhancing antigen production and immune response’ to ‘enhances efficacy’. I personally would have left it as the former title as there is nothing efficacious about these vaccines. If your soul measurement is antibody production, you might come to that conclusion but most antibodies generated post injection never reach the mucosa where the virus enters the body.
The chorus of vaxophiles and mutton minions on twitter were once again out in strong defense of such manufacturing slop because truncated mRNAs lacking a polyA tail will not be translated… at least according to their high school biology understanding of the translation process and lack of understanding of RNAi (RNA interference). They made the assumption that the smaller transcripts seen on Agilent were simply lacking polyA tails. Never assume when you have a sequencer.
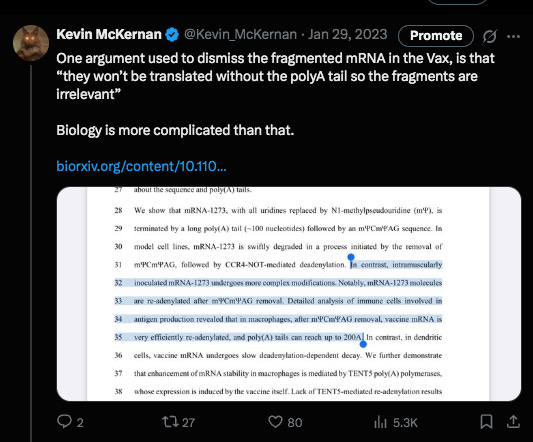
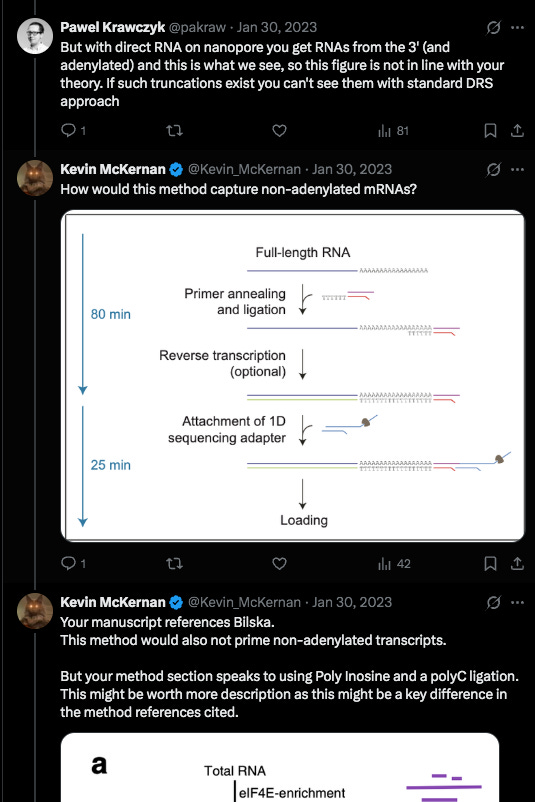
The debate with the author was in regards to if his methods would pick up truncated mRNA that was lacking a polyA tail?
There was some confusion over this as his methods section referenced a paper that couldn’t detect them but these authors modified them in a way where they think they can see them.
The problem I saw in their cited methods was that they first captured the mRNA they wanted to sequencing using a PolyT bead so anything without a PolyA tail would never end in their library leading them to falsely conclude they don’t exist.
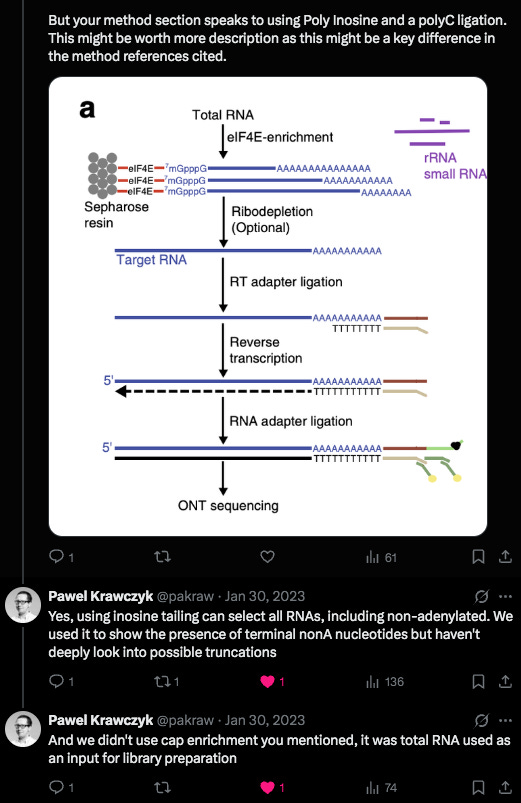
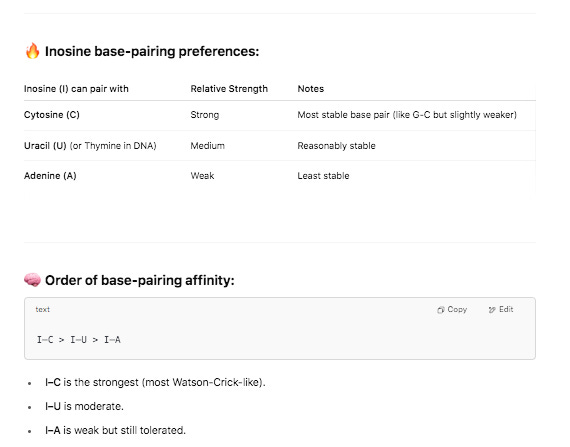
Krawczyk clarified a detail in their methods. This was a clever modification they used. Inosine is a semi-universal base. We used this at the 6th position in our Oligos used for SOLiD sequencing. T4 DNA ligase has very little scrutiny for base pairing fidelity 6 bases away from the ligation junction so you could use a sloppy base and the enzyme didnt care.
But when this sloppy base is at the ligation junction of the enzymes action, it exhibits a strong preference for certain bases. It doesn’t bind G so PolyC primers are often added to complement its affinity for C,T and A. These methods aren’t perfect when you’re asking a polymerase to extend on PolyI and PolyC but they should pick up some truncated transcripts.
Other important details on the methods used to extract this RNA:
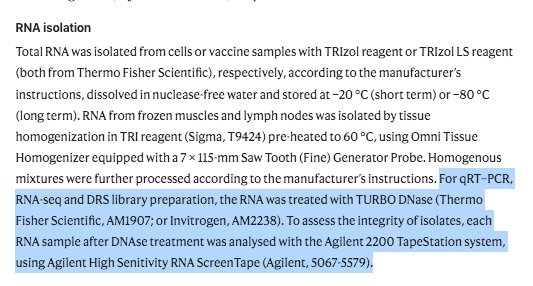
Once again, even though they are using TURBO DNase prior to making these libraries. This should erase most of the DNA present making the plasmid DNA in the Moderna vaccines harder to detect. Nevertheless, we can still find Moderna plasmid DNA in Krawczyk et al Illumina Sequencing data. In absence of this DNaseI treatment the DNA levels would be at even higher coverage levels. This is an important topic given the mRNA longevity assumptions being solely a cause of Re-Adenylation. DNA is known to persist for weeks to months. RNA is hours to days. Modified RNA? We don’t know and we are finding out in real time after vaccinating the human population.
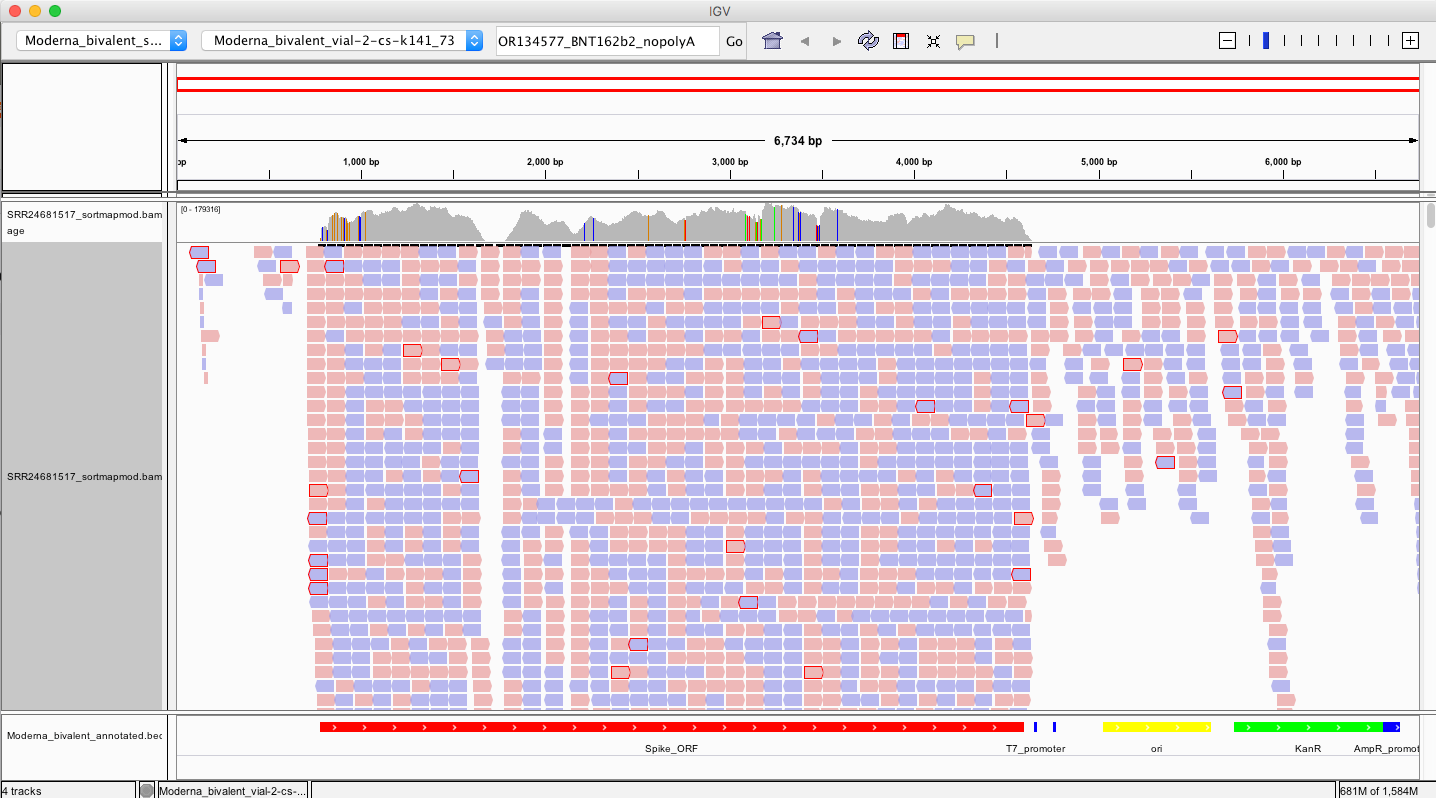
So if we want to find evidence of the 3’ ends of these vaccines having unexpected sequences, we must look at the 3’ end of the modRNA or the left side of that Spike_ORF in red (~790bp).
Recall Transcription runs from the T7 promoter (~4500bp) to the end of the spike on the left.
The reads on that 3’end boundary that are outlined in red are broken paired end reads. That means one 101bp read partially or fully maps to spike and the other read does not. Often times these spike reads are Soft-Clipped… meaning one part of the 101bp read maps to spike and the other part of the read is something else?
What do we see?
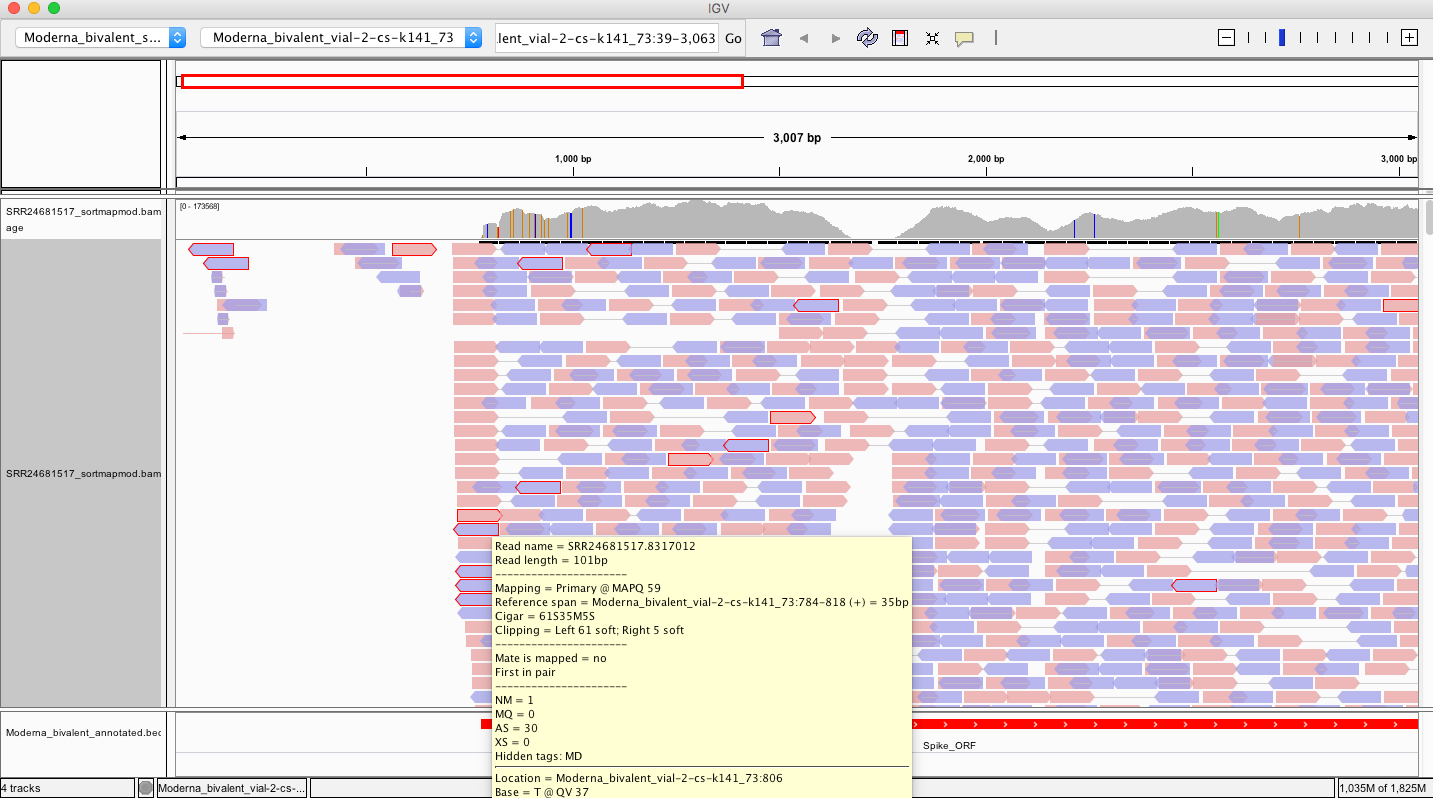
Here is a read where the left 61 bases doesn’t map to spike but 40bp do.
So lets BLAST the 61bp soft clip and see what else is in here.
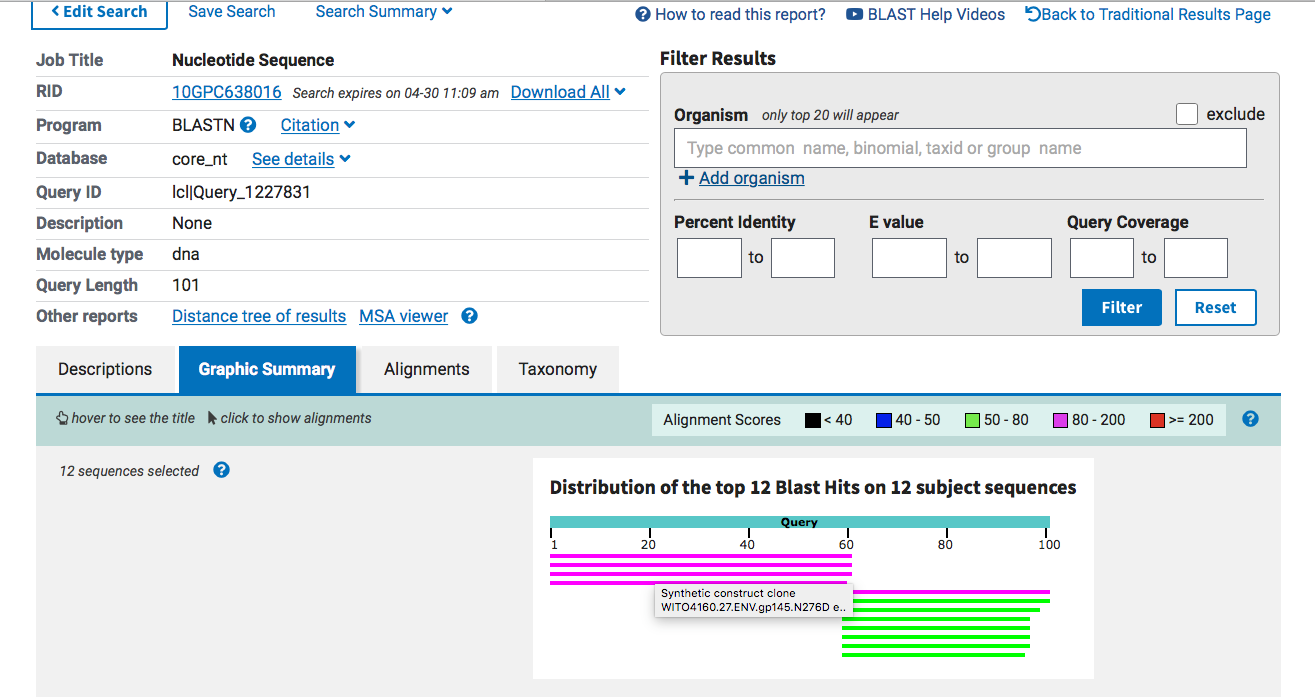
The right side of this BLAST diagram is hitting spike (Green) and the left side is hitting something completely unexpected (Pink).
Green is top BLAST hit and Pink is the second BLAST hit.
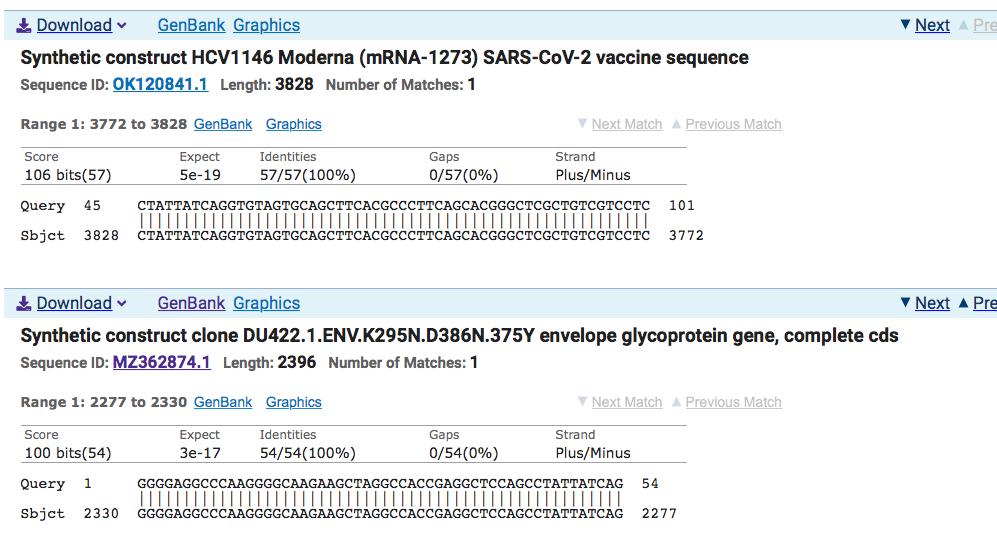
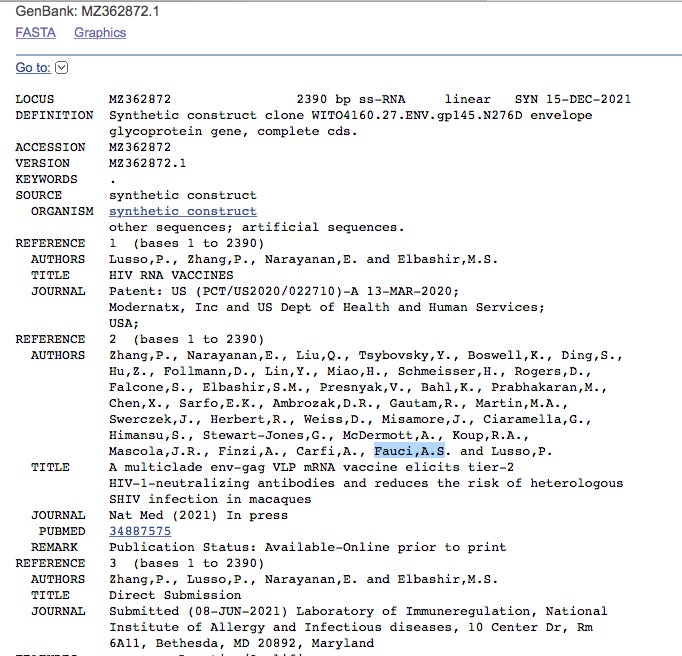
Click on MZ362874.1 and it points to a Moderna HIV vaccine patent filed March 13th, 2020 where Anthony Fauci is an author. The patent was published on Patent: US (PCT/US2020/022710)-A 13-MAR-2020
The NIH makes note of this on their website.
This is supported by more than 1 read with different start points.
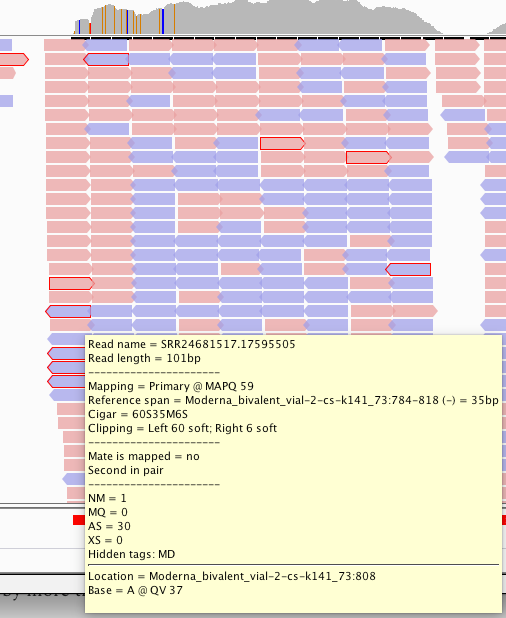
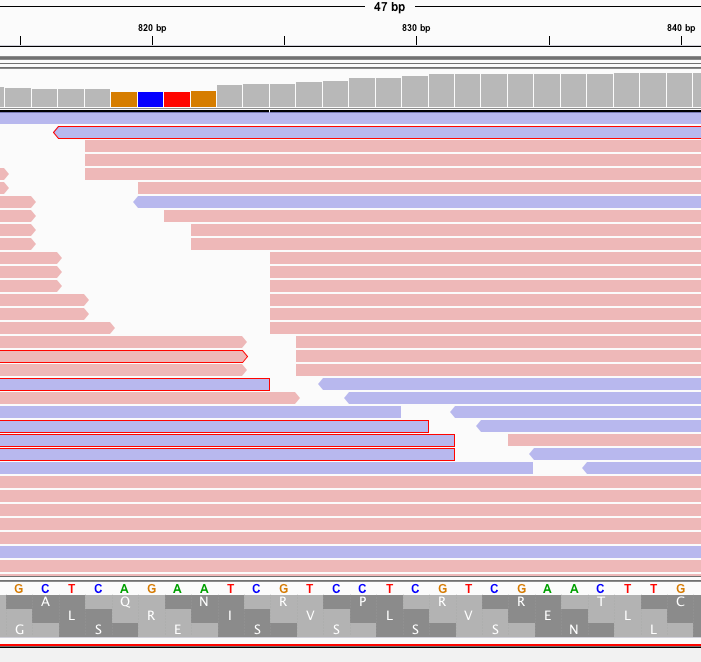
In fact, its supported by a truckload of reads. You’ll note the sequence where the reads disagree with the reference are overlaid in text on the pink and blue reads and do not match the Moderna mRNA1273 vaccine sequence on the bottom track.
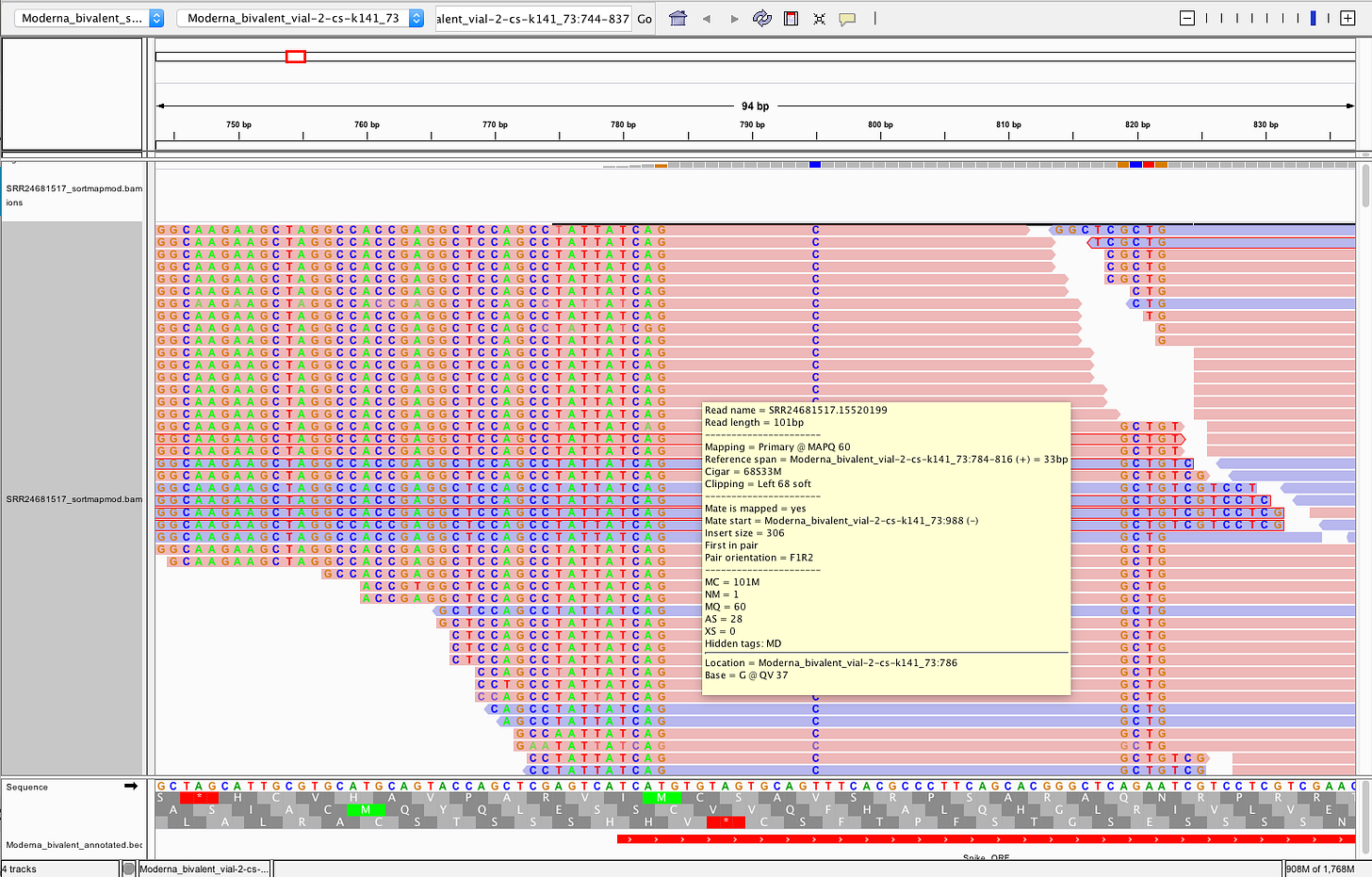
Those non-matching soft-clipped read are chimeric transcripts and they BLAST to this paper.

So how on earth did this get into this SARs-CoV-2 vaccine and what is gp145 doing covalently linked to spike? That doesn’t make any sense unless there is an IRES element between them? IRES = Internal Ribosomal Entry Sites. These enable polycistronic sequences to be translated in parallel from two promoters. Think of IRES an an internal promoter that recruits its own Ribosome for parallel translation of proteins from a single long mRNA. These do not have a consensus sequence we can look for. IRES elements are difficult to predict from sequence alone and require biological validation.
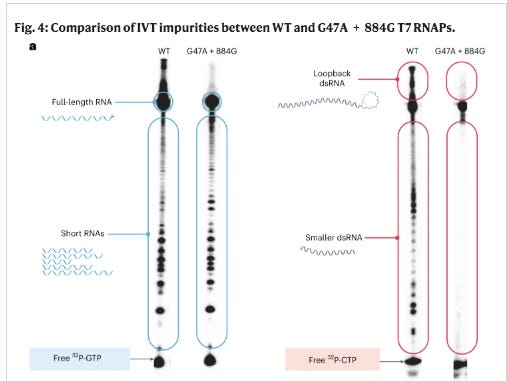
There is another more likely hypothesis given this unexpected sequence is from a Moderna patent. It could be a contaminant plasmid in Moderna’s IVT reaction which template switches and makes chimeric transcripts with two different vaccine targets? Template switching occurs when the T7 Polymerase is transcribing a template and then hops onto another template and makes chimeric RNA. This can happen in cis or in trans. Cis template switching is often called loopback template switching and occur more often when there are palindromic sequences on the RNA (Figure 1A). Trans Template switching occurs when two different DNAs have similar short sequences that terminal end of an RNA can decide to re-prime off two different templates (Figure 1B).

Moderna having the highest GC content in their spike codon optimization was notorious for template switching. They published a paper in 2023 showcasing their mutated T7 polymerase designed to eliminate this artifact. In doing so they lifted the veil on how much of this template switching their normal IVT process generated. See Figure 4 below. Those wildtype (WT) smears are all template switches. The mutated polymerase eliminates this. This is the source of the mess in BlotGate. The Poor RIN numbers and smeary gels; those are template switch artifacts. I just never expected to see another Moderna HIV sequence with Anthony Fauci’s name on it, in the C19 vaccines!

I recall some people claiming to test positive for HIV post C19 vaccination. I don’t know the details of those tests to know if this is playing a role but it has no business being in this vaccine sequence if it in fact was never disclosed (the plasmid DNA wasn’t but this HIV sequence might exist in the sequence they gave the regulators).
Note the authors of Kawczyk have no overlap in name, institution or funding with the Fauci paper. In the appendix, I have tried to comb through ever reagent mentioned in Krawczyk et al to see if this could be an artifact of any other oligo they listed. Using Blastn -task blastn-short I have yet to find one of the oligos listed in their supplementary table could explain it.
A final hypothesis
all Moderna vaccines may share some 3’ sequence and our original assemblies of a Moderna Bivalent Vaccine may have some components missing on the 3’end where the 2 different spike sequences are cloned into the plasmid. When you assemble reads from 2 vaccines, the assemblers try to smash the difference together to draw a consensus. This is could be an artifact of assembling bivalent vaccines and our reference is missing these HIV components. These HIV components in spike are a known controversy since the Pradhan paper.
This final hypothesis is partially supported by mapping this HIV sequence to the monovalent Moderna vaccine from the Fire lab in GitHub. These reads are not public and they did not include the plasmid backbone leaving this as a bit of a guessing game.
I bet if the Fire lab put their reads public, the world would have known about the plasmid contamination 2 years before we ran into. I asked for them but they were never supplied. You can clearly see the DNA plasmid reads in these data and its a bit odd running a plasmid finishing project on sequences that are coming out of animals they shouldn’t be in 4 years into this.

Conclusions
RNA-Seq experiments exploring the fidelity of the 3’ ends of the Moderna Vaccine have given us an interesting insight into a possible mechanism of RNA persistence: Re-adenylation of the messages in-vivo. Inadvertently, the data also demonstrates the plasmid DNA contamination despite methods that should greatly reduce DNA copy number with DNaseI. The data also sheds light on another form of potential contamination in the C19 vaccines; other mRNA vaccines currently in Moderna’s pipeline. With ubiquitous DNA sequencing capacity available to sequence over 9M SARs-CoV-2 genomes, it seems irresponsible for the vaccines to be tolerating RNA of the wrong size in the vaccines. Had these vaccines been sequenced prior to approval, these low RIN scores would be much better understood prior to administering them to the global population.
Appendix-
Vaccine plasmid DNA contamination has now been found in post vaccination of people and mouse models in…
- Chakraborty et al
- Ryan et al
- Odak et al
- Lee et al
- Knabl et al.
- and Now Krawczyk et al.
Please feel free to comment or critique this work. Its a real head scratcher.
Methods
Download the Illumina RNA-Seq reads from NCBI’s SRA.
SRR24681511-SRR24681519 are all from this project. They show varying degrees of this DNA and HIV vaccine contamination. To find the exact reads I’m displaying here, focus on SRR24681517
fastq-dump –split-files SRR24681517
cutadapt -a AGATCGGAAGAGC -A AGATCGGAAGAGC -q 20 -m 20 -o trim_SRR24681517_1.fastq -p trim_SRR24681517_2.fastq SRR24681517_1.fastq SRR24681517_2.fastq
Download this Moderna vaccine reference. Name it OR134578.1
#Format the reference with Seqkit
seqkit seq -w 60 OR134578.1> Moderna_bivalent_seqkit.fa
#Map reads to the formatted Reference Moderna_bivalent_seqkit.fa
bwa mem -t 16 Moderna_bivalent_seqkit.fa trim_SRR24681517_1.fastq trim_SRR24681517_2.fastq |samtools view -@8 -b -F 4 | samtools sort -@8 -o SRR24681517_sortmapmod.bam
#Index the final BAM file for IGV
samtools index SRR24681517_sortmapmod.bam
#Load this into IGV with the reference above.
#If you want to annotate your Reference in IGV, you can Load the below BED file.
Note the header in the Fasta file must have >Moderna_bivalent_vial-2-cs-k141_73 for the below BED file to properly load.

Moderna_bivalent_vial-2-cs-k141_73 779 4595 Spike_ORF 0 + 779 4595 255,0,0
Moderna_bivalent_vial-2-cs-k141_73 4650 4669 T7_promoter 0 + 4650 4669 0,0,255
Moderna_bivalent_vial-2-cs-k141_73 4756 4773 M13_fwd 0 + 4756 4773 0,0,255
Moderna_bivalent_vial-2-cs-k141_73 5025 5614 ori 0 + 5025 5614 255,255,0
Moderna_bivalent_vial-2-cs-k141_73 5735 6545 KanR 0 + 5735 6545 0,255,0
Moderna_bivalent_vial-2-cs-k141_73 6545 6637 AmpR_promoter 0 + 6545 6637 0,0,255
Adaptor sequences for cDNA in Krawczyk. Do any of these account for the off target Moderna/Fauci HIV sequence?
>Moderna/Fauci HIV_Seq
GGGGAGGCCCAAGGGGCAAGAAGCTAGGCCACCGAGGCCACCGAGGCTCCAGCCTATTATCAG
Use
blastn -task blastn-short -db TENT5A-oligo.fa -query Moderna-fauci-HIV-seq.fa
to screen the Moderna-Fauci sequence against all Oligos in their supplement table
An inosine (I)-tailing reaction was performed with 0.5 mM ITP (inosine triphosphate), 1× NEB 2.0 buffer (B7002, NEB) and 2 U poly(U) polymerase (M0337S, NEB) at 37 °C for 45 min, and was terminated by snap-freezing in liquid nitrogen. I-tailed RNA was cleaned twice on KAPA Pure Beads (7983298001, Roche) in a 1× ratio and used for DRS library preparation. I-tailed samples require ligation of a special adaptor RTA_C10, which contains 10 cytosines at the 3′ end: 5′-GAGGCGAGCGGTCAATTTTCCTA AGAGCAAGAAGAAGCCCCCCCCCCCC-3′.
After synthesis of the first cDNA strand, samples from the reverse transcription reaction were pre-amplified in a 12-cycle PCR reaction with RTP and SSPII_Mod_2 (specific for mRNA-1273, with UMI sequence TTTCTGTTGGTGCTGATATTGCTTTVVVVTTVVVVTTVVVVTTVVVVTTTCCACCGACAACACCTTCGTGAGCGG) primers using the LongAmp Hot Start Taq Master Mix.
Other oligos used from their Supplement.
>TaqMan VacProbe2
CACCAAGCTGAACGA
>TaqVac Fw
GATCAGCAACTGCGTGG
>TaqVac Rv
TACACGTTGGTGAAGCAC
>Act_F
CCCAGATCATGTTTGAGACC
>Act_R
ATCACAATGCCTGTGGTACG
>Vac5_fw
CACCTTCAAGTGCTACGG
>Vac5_rv
TAGTTGTAGTCGGCGATCTT
>Gapdh_F
AAGGGCTCATGACCACAGTC
>Gapdh_R
GATGACCTTGCCCACAG
>Tent5a_F
GCTCACTCTCAAGGAGGCTTATG
>Tent5a_R
CTTCAGCTCCACATTTTTGCCAC
>Tent5c_F
CAGTCACCTCCTCTTCCAACG
>Tent5c_R
AACCTGATCCCAGTTGAGCAC
>Hs_ACTB_F
TCACCCACACTGTGCCCATCTACGA
>Hs_ACTB_R
CAGCGGAACCGCTCATTGCCAATGG
>Hs_GAPDH_F
CATGTTCCAATATGATTCCACCC
>Hs_GAPDH_R
CCACGACGTACTCAGCG
>Hs_CHOP_F
TGAGTCATTGCCTTTCTCCTTC
>Hs_CHOP_R
ACATAGGTACCCCCATTTTCATC
>Hs_GRP94_F
TTACTATGCGAGTCAGAAGAAAAC
>Hs_GRP94_R
AAGCATTCTTTCTATTCTATCTCC
>Hs_PERK_F
AAGGTTGGAGACTTTGGGTTAG
>Hs_PERK_R
GAATCTGCTCTGGGCTCATATAC
>Hs_UXBP1_F
CAGACTACGTGCACCTCTGC
>Hs_UXBP1_R
CTGGGTCCAAGTTGTCCAGAAT
>Hs_SXBP1_F
GCTGAGTCCGCAGCAGGT
>Hs_SXBP1_R
CTGGGTCCAAGTTGTCCAGAAT
>Hs_TXBP1_F
TGAAAAACAGAGTAGCAGCTCAGA
>Hs_TXBP1_R
CCCAAGCGCTGTCTTAACTC
>RLucF1
gccatcagattgtgtttgttagtcgctATGATTCCGAGAAGCACGCCGAGAAC
>RLucR1
gcttacggttcactactcacgacgatgTTACTGCTCGTTCTTCAGCACGCG
>RLuc_T7_F2
TAATACGACTCACTATAGGGAGAgccatcagattgtgtttgttagtcgct
>RLuc_A10_R2
TTTTTTTTTTgcttacggttcactactcacgacgatg
>RLuc_A15_R2
TTTTTTTTTTTTTTTgcttacggttcactactcacgacgatg
>RLuc_A30_R2
TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTgcttacggttcactactcacgacgatg
>RLuc_A45_R2
TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTgcttacggttcactactcacgacgatg
>RLuc_A610_R2
TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTgcttacggttcactactcacgacgatg
>RLuc_A90_R2
TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTgcttacggttcactactcacgacgatg
>UTRmod_phi65_GGG_fw
aatatttgtatgTAATACGACTCACTATAGgggaaataagagagaaaagaagagtaag
>UTRmod_phi65_AGG_fw
aatgtttgtatgTAATACGACTCACTATAAgggaaataagagagaaaagaagagtaag
>UTRpf_T7_phi65_GGG_fw
ctagttgtatgTAATACGACTCACTATAGGGAATAAACTAGTATTCTTCTGGTC
>100A_UTRmoderna_rev
ttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttgccgcccactcagac
>100A_pent_UTRmoderna_rev
CTAGAttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttgccgcccactcagac
>pftail_UTRmod TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAGTCATATGCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTgccgcccactcagac
>pftail_pent_UTRmod ctagaTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAGTCATATGCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTgccgcccactcagac
>100A_UTRpfizer_rev
ttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttGCTAGCTCCAGGGTG
>100A_pent_UTRpfizer_rev
CTAGAttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttGCTAGCTCCAGGGTG
>pftail_UTRpf TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAGTCATATGCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGCTAGCTCCAGGGTG
>pftail_pent_UTRpf ctagaTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAGTCATATGCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGCTAGCTCCAGGGTG
>special adaptor
GAGGCGAGCGGTCAATTTTCCTA AGAGCAAGAAGAAGCCCCCCCCCCCC
>SSPII_Mod_2
TTTCTGTTGGTGCTGATATTGCTTTVVVVTTVVVVTTVVVVTTVVVVTTTCCACCGACAACACCTTCGTGAGCGG
Nepetalactone Newsletter is a reader-supported publication. To receive new posts and support my work, consider becoming a free or paid subscriber.
Leave a Reply